A Comprehensive Guide to Protein Acetylation Research: From Modification Discovery to Functional Analysis
A Comprehensive Guide to Protein Acetylation Research: From Modification Discovery to Functional Analysis
In the vast realm of life sciences, protein post-translational modifications (PTMs) have always been a shining research hotspot. The human genome encodes approximately 20,000 proteins, and the diverse array of PTMs acts like magical spells, enabling these proteins to cope with complex and ever-changing life activities. Among them, protein acetylation modification, initially discovered in histones within the cell nucleus, has now emerged in multiple fields such as metabolism, immunity, and autophagy, with its research value becoming increasingly prominent. For research novices, how can they quickly step into the realm of protein acetylation modification research? This article takes an autophagy-related study published in Nature Communications as an example, dismantles its core logic, and presents a clear research framework.
I. Anchoring the Starting Point: Confirming the "Acetylation Identity" of the Target Protein
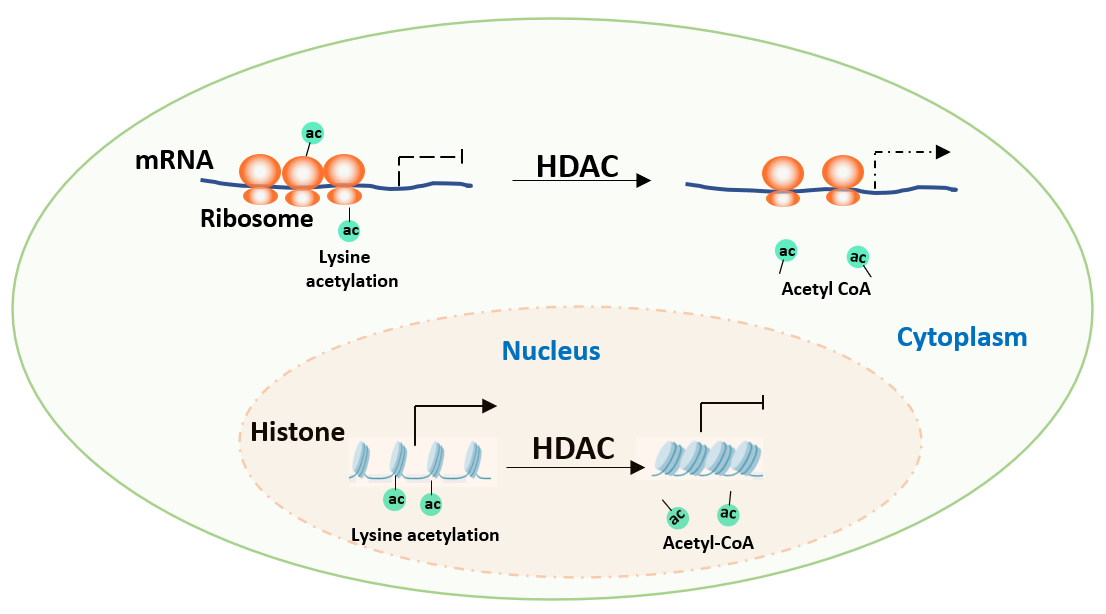
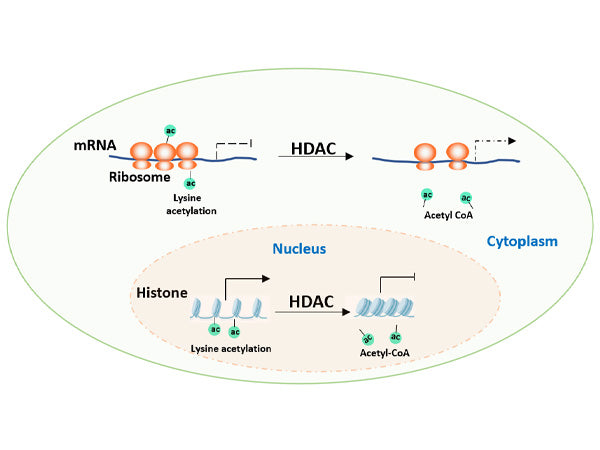
Research begins with exploring whether the target protein undergoes acetylation modification. Taking the autophagy receptor protein p62 as an example, researchers conducted studies using two deacetylase inhibitors: TSA (an inhibitor of the HDAC family) and NAM (an inhibitor of the Sirtuin family). Experiments showed that TSA treatment significantly increased the acetylation level of p62, while NAM had no obvious effect. This difference is crucial, not only confirming the existence of acetylation modification on p62 but also initially suggesting that its deacetylation process is mediated by HDACs rather than Sirtuins, pointing the way for subsequent research.
This step is like a detective determining key clues in a case. Only by clearly identifying the "acetylation identity" of the target protein can further in-depth exploration be initiated.
II. Exploring Regulation: Unlocking the Dynamic Code of Modification Levels
Finding unregulated protein modifications is like searching for a needle in a haystack, while exploring the regulatory patterns of acetylation levels is a key aspect of the research. Researchers treated cells with various stimuli, including starvation and the proteasome inhibitor MG132, and found that only starvation conditions could significantly increase the acetylation level of p62. This result reveals the regulatory role of environmental factors on acetylation modification and also validates the scientific consensus that "regulated modifications are meaningful."
The discovery of regulatory patterns is like finding a key that opens the door to modification mechanism research, allowing researchers to delve deeper into the changing patterns of modifications under specific physiological conditions.
III. Enzymatic Analysis: Locating the "Writers" and "Erasers" of Acetylation
In mammalian cells, acetylation transferases and deacetylases are the core players in regulating acetylation modification. Acetylation transferases such as p300, CBP, PCAF, GCN5, and TIP60 act as "writers," responsible for adding acetylation tags to proteins, while deacetylases such as the HDAC family function as "erasers," removing these tags.
Researchers overexpressed or knocked down different acetyltransferases in cells and found that the overexpression of TIP60 significantly increased the acetylation level of p62, thereby identifying TIP60 as the acetylation transferase for p62. In the screening of deacetylases, based on the earlier hint from TSA treatment, they focused on the HDAC family and found that the overexpression of HDAC6 would decrease the acetylation level of p62, and in vitro experiments further confirmed that HDAC6 could directly remove the acetylation modification of p62.
This process is like identifying the real culprit among numerous suspects, clarifying the key enzymes that regulate modifications and laying the foundation for analyzing the modification mechanism.
IV. Site Identification: Deciphering the Precise "Codons" of Acetylation
Determining the acetylation sites is a crucial step for in-depth research. There are two common methods: one is to purify the target protein in cells for mass spectrometry identification, and the other is to perform mass spectrometry analysis after reacting the purified target protein with acetyltransferases in vitro. Regardless of which method is used, verification through the construction of mutants is necessary — mutating lysine (K) to arginine (R) to simulate the deacetylated state and to glutamine (Q) to simulate the acetylated state.
In the study of p62, through mass spectrometry analysis combined with mutant experiments, its acetylation sites were mainly identified at specific lysine residues in the UBA domain that mediates ubiquitin binding, providing precise targets for subsequent functional research.
V. Functional Analysis: Elucidating the Impact of Modifications on Proteins and Their Biological Significance
After clarifying the acetylation sites, it is necessary to further explore the impact of modifications on the function of the target protein. The acetylation sites of p62 are located in the UBA domain. Researchers constructed mutants that simulate acetylated/deacetylated states and found that acetylation enhanced the binding ability of p62 to ubiquitin and the formation of UBA domain dimers, which is crucial for p62's recruitment of ubiquitinated proteins during autophagy.
At the level of biological function research, combined with p62's role in autophagy, researchers observed at the cellular and animal levels that the acetylation modification of p62 affects the degradation of ubiquitinated proteins and cell viability, thereby revealing its important role under nutritional stress conditions.
Research Framework Summary: Five Steps to Build an Acetylation Modification Research System
- Modification Existence Verification: Determine whether the target protein undergoes acetylation through methods such as inhibitor treatment and immunoprecipitation.
- Regulatory Factor Screening: Clarify the dynamic changes in modification levels using different stimulation conditions.
- Enzymatic Mechanism Analysis: Screen acetyltransferases and deacetylases to identify the key enzymes regulating modifications.
- Site Precise Localization: Identify specific acetylation sites by combining mass spectrometry analysis with mutant experiments.
- Functional In-depth Exploration: Analyze the impact of modifications on the target protein and biological processes from the molecular, cellular to animal levels.
Protein acetylation modification research is like a precise jigsaw puzzle game, with each step being an indispensable puzzle piece. From the initial confirmation of modification existence to the final elucidation of biological functions, each step requires rigorous experimental design and ingenious logical reasoning. For researchers, this research framework is not only applicable to specific proteins like p62 but also serves as a universal key to initiating acetylation modification research. With the advancement of technology and the deepening of research, protein acetylation modification will showcase its unique value in more fields, providing new perspectives and strategies for revealing the mysteries of life and disease mechanisms.
| Product Information | |||||
| Catalog Number | Product Name | Species | Expression System | Conjugation | Price |
| UA010560 | DNAM-1/CD226 Fc Chimera Protein, Human | Human | HEK293 | Unconjugated | $460 |
| UA010326 | Biotinylated DNAM-1/CD226 His&Avi Tag Protein, Human | Human | HEK293 | Biotin | $520 |
| UA010072 | DNAM-1/CD226 His Tag Protein, Human | Human | HEK293 | Unconjugated | $460 |
| UA01001C-B | 重组蛋白 | Inquiry | |||