Proximity Ligation Assay (PLA) Combined with Post-Translational Modifications (PTMs) Studies: A Novel Approach for Protein Research.

Recent Advances
Post-translational modifications (PTMs) refer to chemical alterations that occur after protein synthesis. These modifications can alter the structure, function, and localization of proteins, playing critical roles in cellular signaling, metabolism, and the development and progression of diseases. The Proximity Ligation Assay (PLA), developed by Navinci in Sweden, provides a highly sensitive and specific method for studying PTMs.
Post-Translational Modifications (PTMs)
Proteins are the primary executors of cellular functions, and post-translational modifications finely regulate protein activity, localization, stability, and interactions with other molecules by adding or removing specific chemical groups. Common PTMs include:
-
Phosphorylation: The addition of phosphate groups, typically catalyzed by protein kinases, can alter protein activity, localization, and interactions with other proteins.
-
Glycosylation: The addition of sugar groups, which can affect protein stability, solubility, and interactions with other proteins.
-
Ubiquitination: The addition of ubiquitin proteins, which can mark proteins for degradation.
-
Acetylation: The addition of acetyl groups, which can influence protein activity, stability, and interactions with other proteins.
PTMs play crucial roles in cellular signaling, metabolism, cell cycle regulation, immune responses, and disease development. For example, phosphorylation can regulate enzyme activity, glycosylation can affect protein stability and localization, and ubiquitination can mark proteins for degradation. Abnormal PTMs are associated with diseases; for instance, excessive phosphorylation of certain oncogenes can lead to uncontrolled cell growth, while excessive ubiquitination of tumor suppressor genes can promote tumor progression.
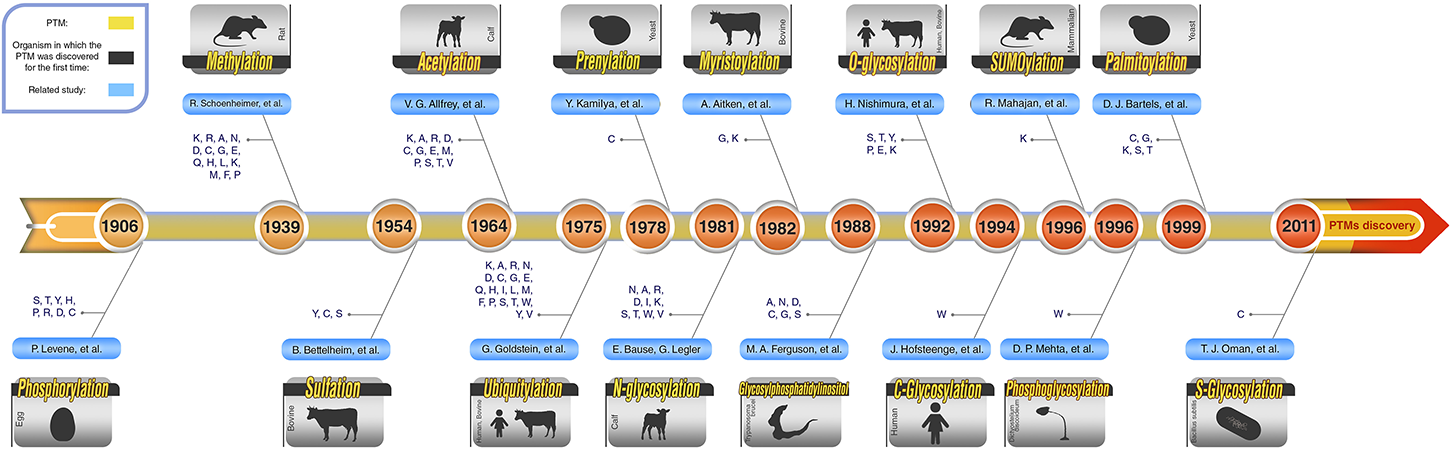
Schematic PTM Discovery Timeline for 10 Major PTMs
Proximity Ligation Assay (PLA)
The PLA technology was developed by Navinci in Sweden. Its core principle is based on signal amplification triggered by proximity effects (Söderberg, O. et al. (2006). Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature Methods, 3(12), 995 - 1000).
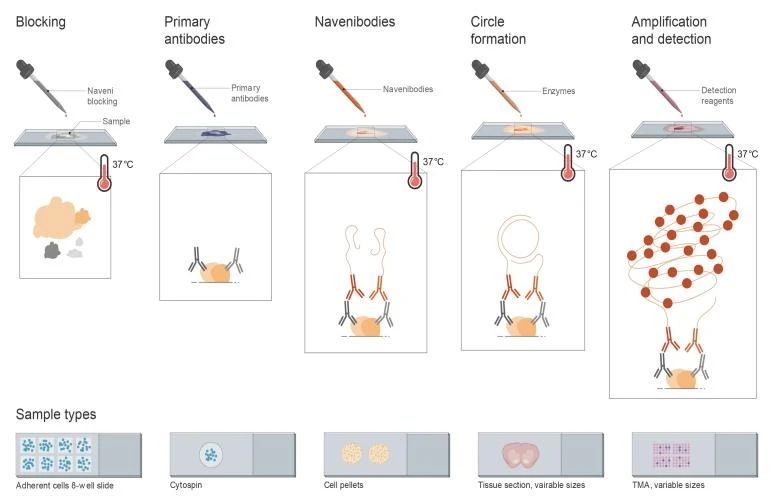
PLA Experimental Workflow:The technology uses two pairs of antibodies, each pair conjugated to a unique oligonucleotide sequence. When these antibody pairs specifically bind to the target protein complex in close proximity (typically within 40 nm), the oligonucleotide sequences attached to the antibodies also come into close proximity. At this point, a DNA ligase connects the adjacent oligonucleotides, forming a complete, amplifiable DNA circle. Subsequently, rolling circle amplification (RCA) is used to amplify this DNA circle, generating a long single-stranded DNA containing multiple repeating sequence units. Finally, fluorescently labeled complementary oligonucleotide probes hybridize to the amplified DNA strand, and fluorescence microscopy or other fluorescence detection equipment is used to detect the fluorescent signal. The intensity of the fluorescent signal is proportional to the quantity of the target protein complex, enabling qualitative and quantitative detection of the target protein complex.
PLA Technology for PTM Detection: Application Examples
Phosphorylated HER2 Detection
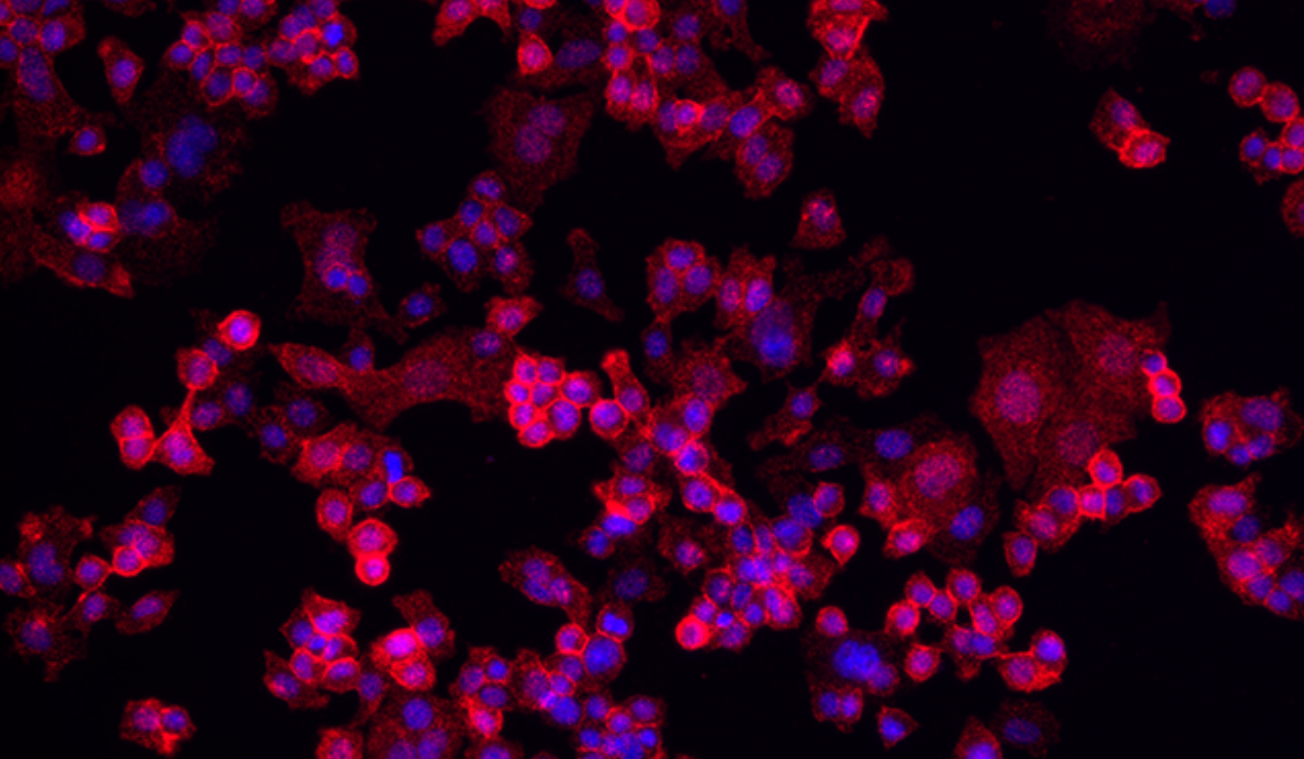
Using the Naveni pY HER2 kit, detection fluorophore TEX615.
Phosphorylated EGFR Detection
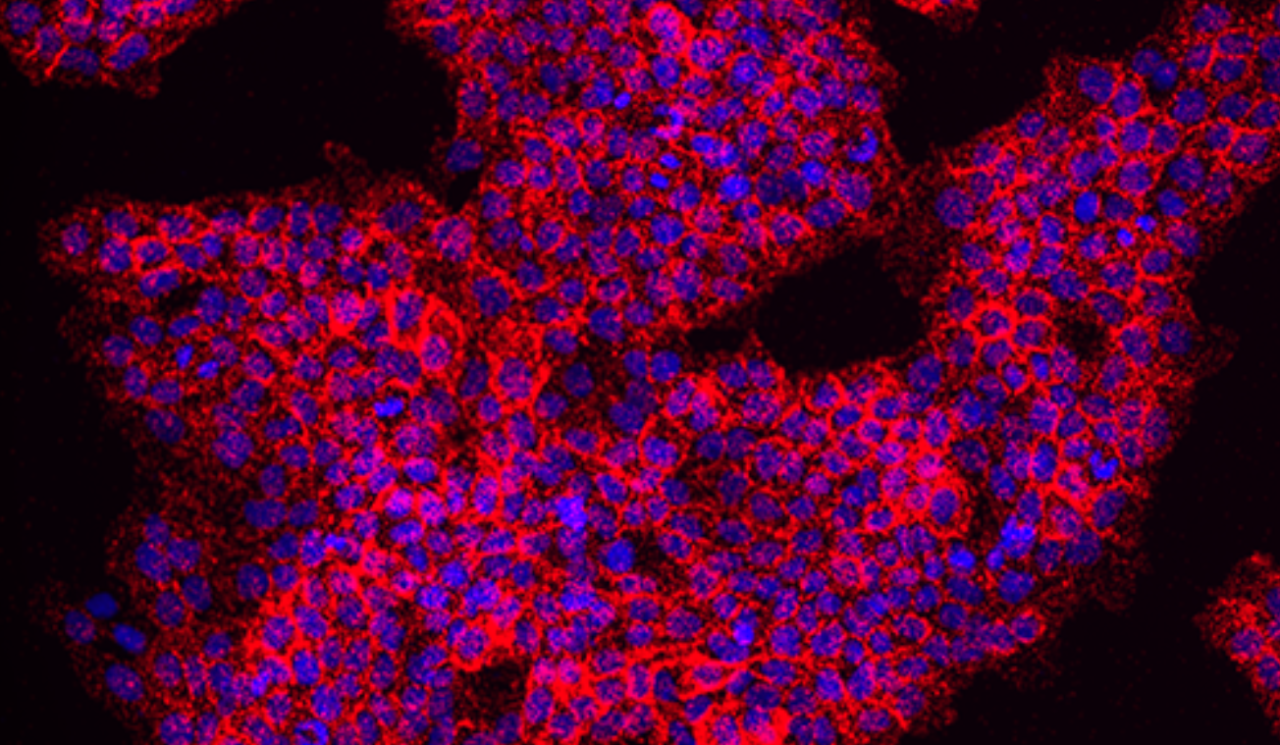
Using the Naveni pY EGFR kit, detection fluorophore TEX615.
Additionally, START's range of modification-specific antibodies can be paired with the NaveniFlex kit to detect various phosphorylated proteins.
Product Information
| Gatalog Num | Product Name | Product Parameters | Price |
| S0B0748 | Phospho-Stat1 (Tyr701) Recombinant Rabbit mAb (S-601-78) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0631 | Phospho-IκBα (Ser32/36) Recombinant Rabbit mAb (S-751-40) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0611 | Phospho-Akt (Ser473) Recombinant Rabbit mAb (S-622-64) | Host : Rabbit | $100 |
| Conjugation : Unconjugated | |||
| S0B0597 | Phospho-mTOR (Ser2448) Recombinant Rabbit mAb (S-705-7) | Host : Rabbit | $100 |
| Conjugation : Unconjugated | |||
| S0B0596 | Phospho-Akt (Ser473) Recombinant Rabbit mAb (S-763-7) | Host : Rabbit | $100 |
| Conjugation : Unconjugated | |||
| S0B0518 | Phospho-GSK-3β (Ser9) Recombinant Rabbit mAb (S-748-20) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0363 | Phospho-Akt (Ser473) Recombinant Rabbit mAb (S-510-64) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0282 | Phospho-S6 Ribosomal Protein (Ser235/236) Recombinant Rabbit mAb (S-R203) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0257 | Phospho-IκBα (Ser36) Recombinant Rabbit mAb (S-R102) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0087 | Ubiquitin Recombinant Rabbit mAb (SDT-R095) | Host : Rabbit | $100 |
| S0B2355 | EGFR (L858R) Recombinant Rabbit mAb (SDT-421-202) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated | |||
| S0B0319 | Phosphotyrosine Recombinant Rabbit mAb (S-R207) | Host : Rabbit | Inquiry |
| Conjugation : Unconjugated |