Product Specification
| Species |
Bovine Pancreatic |
| Synonyms |
DNASE,Deoxyribonuclease-1 |
| Expression System |
E.coli |
| Molecular Weight |
72kDa (Reducing) |
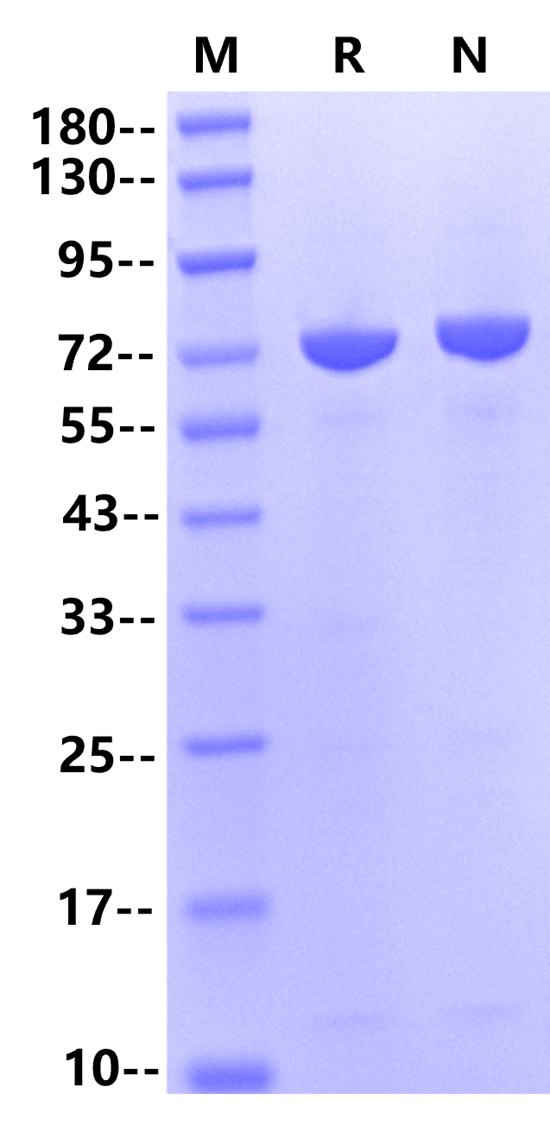
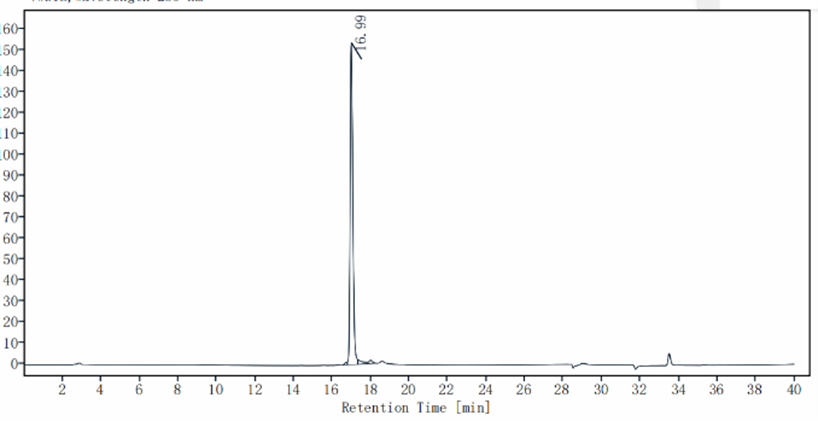
| Purity |
>95% by SDS-PAGE&HPLC |
| Conjugation |
Unconjugated |
| Tag |
His Tag, MBP Tag |
| Physical Appearance |
Liquid |
| Storage Buffer |
10 mM Tris-HCl, 2 mM CaCl2 ,50% Glycerol,(pH 7.6, 25°C) |
| Stability & Storage |
Store at -25 ~ -15℃
for 2 years
|
| Reference |
[1] Vanecko S, Laskowski M. Studies of the
Specificity of Deoxyribonuclease I[J]. Journal of Biological Chemistry, 1961,
236(236):3312-6.
[2] Kienzle N, Young D, Zehntner S, et al. DNaseI
treatment is a prerequisite for the amplification of cDNA from episomal-based
genes[J]. Biotechniques, 1996, 20(4):612-6.
[3] Michael,R, Green, et
al. Human β-globin pre-mRNA synthesized in vitro is accurately spliced in
xenopus oocyte nuclei[J].Cell, 1983, 32(3):681-694.
|
Background
DNase I
(Deoxyribonuclease I), can digest single or double-stranded DNA to produce mono
deoxynucleotides or single or double-stranded oligo deoxynucleotides, its
optimal working pH range is 7-8. DNase I activity is dependent on Ca2+
and can be activated by other bivalent metal ions such as Mg2+, Mn2+,
Zn2+, etc. In the presence of Mg2+, the enzyme can
randomly recognize and cut any site on any strand of DNA. In the presence of Mn2+,
two strands of DNA can be cut at the same site to form sticky ends with flat
ends or 1-2 nucleotides protruding.
Components
Storage
Solution: 2 U/ul DnaseⅠ、10mM Tris-Hcl、2mM CaCl2、50%Glycerol (pH7.6, 25℃)
10*Reaction
Buffer: 100mM Tris-Hcl、25mM MgCl2、5mM CaCl2 (pH7.6, 25℃)
Protocol
This step is suitable
for linearization of 1 μg DNA (≥100 nt) and can be scaled up according to
experimental needs.
1)Add the following components in
sequence
|
Components
|
Volume
|
Plasmid DNA
|
1μg DNA
|
10*Reaction Buffer
|
2μl
|
DnaseⅠ (2U/μl)
|
1μl
|
RNase-free ddH2O
|
Up to 50μl
|
2)Incubate at 37°C 1 h.
Guidelines
1. EDTA should be added to a final concentration of 5 mM to protect RNA from being degraded during enzyme inactivation 2. Please avoid repeated freeze-thaw cycles
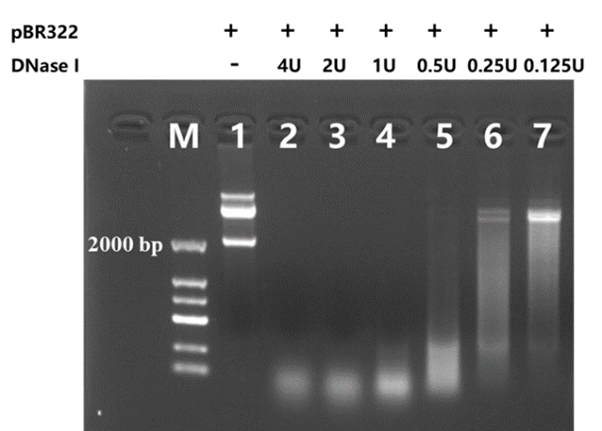
Unit Definition
One unit is defined as the amount of enzyme which will completely degrade 1 µg of pBR322 DNA in 10 minutes at 37°C in DNase I Reaction Buffer.