Aiding the Exploration of the "Second Code" of Life Information: Epigenetics Solutions
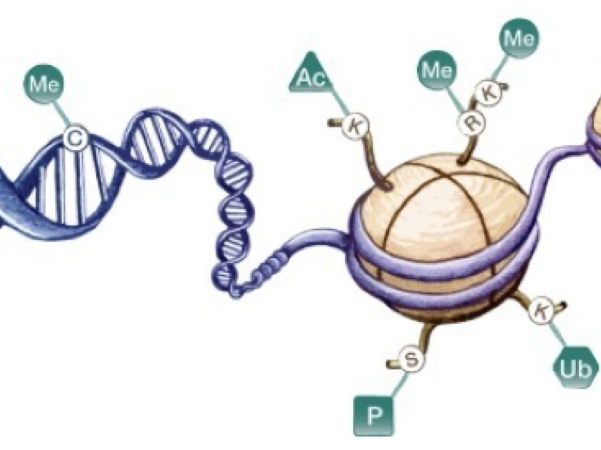
I. From "Predestined" to "Acquired Modification": Genetics Beyond DNA Sequences
Classical genetics regards DNA as an unalterable "blueprint". However, phenotypic differences between monozygotic twins and distinct functions of different tissue cells suggest the existence of a set of reversible information "written above DNA" — Epigenetics [1]. It determines when, where, and at what intensity genes are "read" through three core mechanisms, forming the "second code" of life.
II. In-depth Interpretation of Three Core Mechanisms
1. DNA Methylation: The "Roadblock" of Gene Expression
Adding a methyl group (-CH₃) to CpG sites can physically block the binding of transcription factors, thereby silencing genes. This "roadblock" is added by DNMT and removed by TET, dynamically regulating embryonic development, cell differentiation, and tumorigenesis [2][3]. In 2025, large-scale population methylome studies further confirmed that Differentially Methylated Regions (DMRs) are common targets for early cancer screening and stress-resistant crop breeding.
Key Research Directions and Indicators:
• Maintenance Methylation vs. De Novo Methylation: DNMT1 is responsible for post-replication maintenance; DNMT3A/3B are responsible for de novo methylation establishment.
• Hemimethylation: Hemimethylated CpG generated after replication serves as an intermediate indicator for measuring methylation homeostasis.
• Hydroxymethylation (5hmC): TET enzymes oxidize 5mC to produce 5hmC, which is regarded as the "entry point for demethylation" and its content can be used as a pluripotency marker of stem cells [3].
• Methylation Heterogeneity: The "methylation distance" within tumors can predict immunotherapy response [4].
Absin Products:
|
Catalog No. |
Product Name |
Application |
|
Chromatin Immunoprecipitation (ChIP) Kit |
Enrichment of methylated-CpG binding protein MeCP2 to verify methylation-transcription coupling |
|
|
Human DNMT1 ELISA Kit |
Detection of the content of key DNA methylation enzyme |
|
|
Human DNMT3A ELISA Kit |
Detection of the content of key DNA methylation enzyme |
2. Histone Modification: The "Rheostat" of Chromatin
Acetylation, methylation, lactylation, propionylation, etc., on histone tails can alter chromatin compactness, forming the "histone code" [5].
Main Modification Types and Functions:
• Metabolism-Epigenetics Crosstalk: Metabolites such as lactate, succinyl-CoA, and β-hydroxybutyrate act as "acyl donors" to directly regulate gene expression [6].
• "Broad" H3K4me3: Covers the entire gene body in the early embryonic stage, maintaining genome silencing until zygote activation [7].
• Histone Phosphorylation: γH2AX (S139ph) serves as a "distress signal" to recruit repair factors when DNA is damaged [8].
Absin Products:
|
Catalog No. |
Product Name |
Application |
|
abs145130 |
Rabbit anti-Acetyl-Histone H3 (Lys56) Monoclonal Antibody (R02-5J8) |
Antibody for detecting acetylated Histone H3 |
|
abs145132 |
Mouse anti-Acetyl-Histone H3 (Lys9) Monoclonal Antibody |
Antibody for detecting acetylated Histone H3 |
|
abs500002 |
Animal Histone Extraction Kit |
Histone extraction |
3. Non-Coding RNA (ncRNA): The "Commander" of Gene Networks
ncRNAs (miRNA, lncRNA, circRNA) globally regulate genes at the post-transcriptional or chromatin level through base pairing or recruitment of protein complexes; RNA itself can carry more than 160 types of chemical modifications, forming the "epitranscriptome".
Research Directions:
• RNA Modification-Metabolism Coupling: Hyperglycemia induces high m6A methylation, accelerating the degradation of mRNA of vascular endothelial inflammatory factors [9].
• lncRNA-Guided Modification: lncRNA HOTAIR recruits PRC2 to trigger H3K27me3-mediated silencing; meanwhile, it is modified by m6A to regulate its stability [10].
• circRNA "Sponge" Mechanism: circRNA evades immune recognition through m6A modification, continuously sequestering miRNAs and affecting the tumor immune microenvironment [11].
Absin Products:
|
Catalog No. |
Product Name |
Application |
|
abs60260 |
Blood miRNA Extraction Kit |
miRNA extraction |
|
abs60261 |
Tissue miRNA Extraction Kit |
miRNA extraction |
|
abs60262 |
Cell miRNA Extraction Kit |
miRNA extraction |
|
abs60264 |
Plasma/Serum miRNA Extraction Kit |
miRNA extraction |
|
abs60252 |
Small Nucleic Acid Transfection Reagent |
Transfection of nucleic acids within 200bp, such as siRNA, miRNA mimics, miRNA inhibitor |
|
abs60265 |
miRNA Polyadenylation Reverse Transcription Kit |
miRNA reverse transcription |
|
abs60271 |
miRNA Polyadenylation Dye-Based Fluorescence Quantitative PCR Kit |
miRNA reverse transcription + qPCR |
|
abs60341 |
Dual-Luciferase Reporter Gene Assay Kit |
Multicomponent dual-luciferase reporter gene detection |
|
abs60676 |
DUG-LTM Dual-Luciferase Reporter Gene Assay Kit |
Single-component dual-luciferase reporter gene detection |
III. Technological Revolution: Six Hot Trends in 2025
① Single-Cell Epigenomics | scATAC-seq, scChIP-seq | First mapping of the complete epigenetic roadmap of human embryos at day 7 [12]
② Spatial Epigenomics | Spatial-CUT&Tag, MERFISH | Lactylation gradient in tumor-margin-normal three regions of colorectal cancer [13]
③ 3D Genomics | Micro-C, Hi-C 2.0 | Discovery of enhancer "phase-separated droplets" regulating TAD boundaries [14]
④ Epigenetic Clock | WGBS + Machine Learning | Blood cfDNA methylation clock predicts cardiovascular events within 5 years (AUC=0.92) [15]
⑤ Epigenetic Editing Therapy | dCas9-TET, CRISPRoff | First in vivo activation of the silenced FMR1 gene, with safe Phase I clinical trial [16]
⑥ Crop Epigenetic Breeding | RRBS, ATAC-seq | Lactylation modification of wheat vernalization gene VRN1 determines flowering time [17]
References:
[1] Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012.
[2] Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018.
[3] Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013.
[4] Rauschert S. et al. Maternal Smoking During Pregnancy Induces Persistent Epigenetic Changes. Front Genet. 2019.
[5] Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000.
[6] Zhang D. et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019.
[7] Liu Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk. Nat Biotechnol. 2013.
[8] Bonaldi T. et al. Monocytic cells hyperacetylate chromatin protein HMGB1. EMBO J. 2003.
[9] Datta J. et al. A new class of quinoline-based DNA hypomethylating agents. Cancer Res. 2009.
[10] Gomez J.A. et al. The lncRNA Nest controls microbial susceptibility and epigenetic activation. Cell. 2013.
[11] Memczak S. et al. Circular RNAs are a large class of animal RNAs. Nature. 2013.
[12] Zenk F. et al. Single-cell epigenomic reconstruction of developmental trajectories. Nat Neurosci. 2024.
[13] Liu C. et al. Spatial-CUT&Tag maps spatial epigenomic profiles. Nat Methods. 2022.
[14] Krietenstein N. et al. Ultrastructural details of mammalian chromosome architecture. Nature. 2020.
[15] Horvath S. DNA methylation age of human tissues. Genome Biol. 2013.
[16] Nuñez J.K. et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2024.
[17] Niu D. et al. A molecular mechanism for embryonic resetting of winter memory. Nat Plants. 2024.




